Protein Structure Databases: Protein Data bank (PDB), PDBe & PDBsum
The Protein Data Bank (PDB), PDB Europe (PDBe), and PDBsum are protein databases that archive and present structural and functional data on proteins, protein complexes, and other biological macromolecules. In this section, we discuss some historical perspectives and the main features of each database.
There are numerous resources related to structural bioinformatics available online. Some are general, while others focus on specific aspects of proteins, protein families, and functions, among other topics. Here, we highlight the databases commonly used and essential for structural biology and structural bioinformatics: the RSCB Protein Data Bank (PDB) and two affiliated European versions, PDBe and PDBsum. PDB and PDBe are members of the Worldwide PDB (wwPDB) organization, which also includes the Biological Magnetic Resonance Data Bank (BMRB), the Electron Microscopy Data Bank (EMDB), and PDB Japan (PDBj).
The PDB History
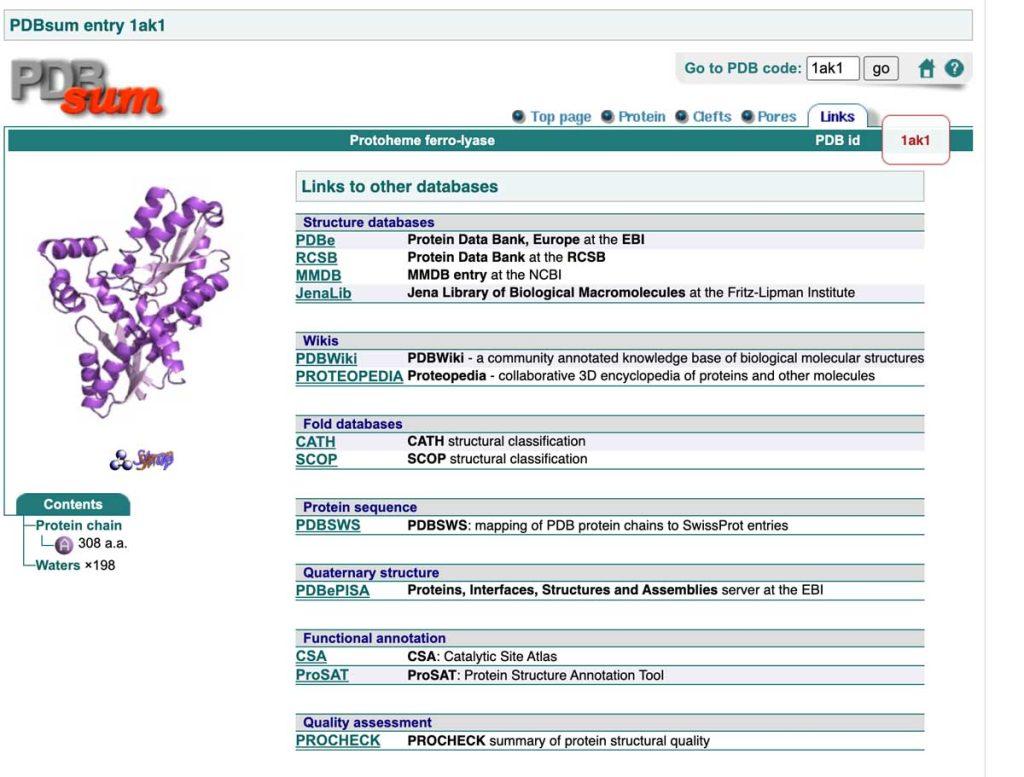
For an extended period, the primary database for protein structures was the RCSB Protein Data Bank, established at the beginning of the 1970s. Only a few structures existed when it was created. The Internet had not yet been invented, and no online databases existed. Until the early 1990s, data access required sending a request to the PDB, which would then send magnetic tapes containing the available structures. These tapes were mounted on tape stations to transfer the files with the atomic coordinates of the structures to a computer. At that time, PCs did not exist, and computers generally did not have graphics cards. Expensive mainframe computers were connected to terminals and separate graphic stations for visualizing the three-dimensional structure. The whole setup was costly, and academic departments could afford only one or two such stations. People had to make reservations to use the system! And we also need to mention that protein X-ray crystallography was the only method for structure determination, and the number of structures in the data bank was limited. As we can see from the image above, starting from the 1990s, things changed, and the PDB content growth rate started to increase rapidly.
There are several reasons for this structural revolution. Before the cloning era, proteins were purified directly from cells, which substantially limited availability − there is always a limited number of copies of a particular protein in a cell. When cloning techniques started to enter the lab, the number of different proteins and their quantity available for crystallization increased drastically. Another essential factor was the introduction of synchrotron radiation for X-ray data collection. Several synchrotrons worldwide currently provide high intensity X-rays for quality X-ray data collection. In addition, synchrotrons reduced the time required to optimize crystallization conditions since much smaller crystals can be used at synchrotrons. In the early days of crystallography, crystallization conditions had to be optimized to grow crystals large enough for the relatively low-intensity laboratory X-ray sources. An important factor was also the introduction of low-cost personal computers with ever-increasing computational and graphics processing power. Cheaper computers also meant new software, which became much more user-friendly. Then came the structural genomics era – large consortia were formed to develop new crystallization technologies and solve large numbers of protein structures. With the increasing number of solved structures, new tools for analyzing protein sequences and structures were rapidly developed, and the number of protein databases increased.
The Protein Data Bank
Although the number of structures in the PDB is rapidly increasing, one should remember that PDB entries are not unique; there are often many entries for the same protein in the database – variants containing mutations, complexes with various ligands (substrate analogs, inhibitors, cofactors), or complexes with other proteins. This can lead to confusion when we try to retrieve a structure from the PDB. Which one should you choose if multiple entries for the same protein exist? For our purposes, we also need to remember that not all structures in the PDB are created equal when it comes to structure quality. We must know how to identify the best-quality structure!
Currently, by using the PDB on the Internet, we can easily find the structure of the protein of interest and evaluate its quality. All we need to do is type the protein’s name into the search field. Typically, the output yields many hits, some of which may be unrelated to the search query. PDBsum and PDBe (PDB Europe) usually provide more refined search results. Refining the search using the options available on the PDB site is also possible.
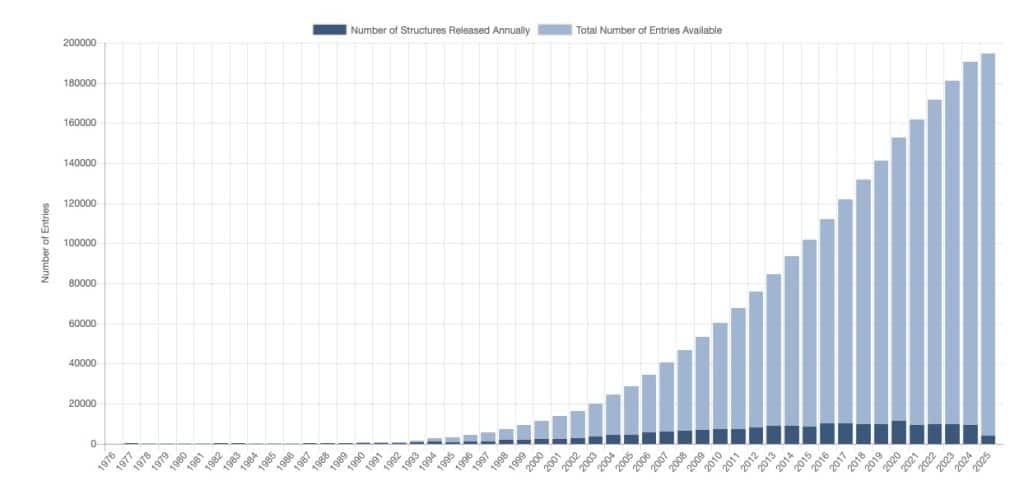
All databases, such as PDB, PDBe, and PDBsum, typically offer many additional links where more information can be found. Below is an example from the PDBsum link page (for mobile view, please click here).
PDB Europe (PDBe)
The PDBsum Database
The PDBsum database…